New! ASH is APA-based, full-length-enriched, single-stranded cDNA subtraction hybridization library.
This unique offering will be offered at an introductory price
for a limited time. Please inquire for more details.
Also offering a reverse subtraction library for an additional fee. Please contact us to inquire.
ASH library specifications
- Insert sizes of about 1200bp (based on colony PCR_ with 10% of clones <500bp, and a >95% recombinant rate.
- Number of clones: >30,000 for a subtracted cDNA library. Additional clone available upon request for an extra fee.
- Full-length enriched.
- Directional cloning: Subtracted cDNA will be ligated directionally in pBluescript-directional cloning vector modified by Bio S&T.The direction will be 5’-end/EcoRI and 3’-end/XhoI.
- Libraries will not be arrayed nor amplified. Bio S&T will provide transformed cells.
- See below Figure 1:
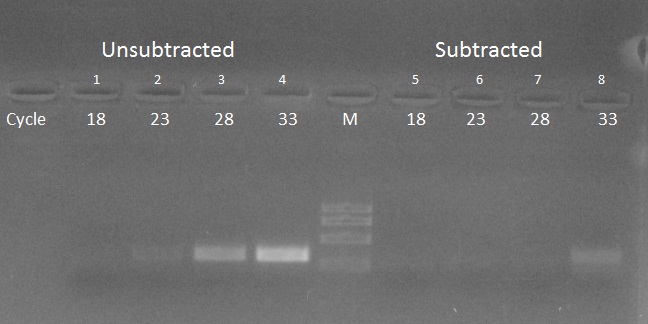
Figure 1: ASH cDNA - Actin gene reduction in grapefruit.
Tested and driver cDNA was prepared from grapefruit citrus leaves. PCR was performed on the unsubtracted cDNA (lanes 1-4) and subtracted cDNA (lanes 5-8) using the actin gene. Note the important reduction of the abundant actin gene.
ASH library construction steps
- Extraction and purification of total RNA.
- Anti-sense strand synthesis of tester (Full-length enriched).
- Sense strand synthesis of driver by APATM (full-length enriched) and labeling.
- Hybridization at a ratio of >500 :1 (driver: tester).
- Removal of double-stranded hybrids.
- PCR amplification of remaining single-stranded tester cDNA.
- Purification and enzyme digestion, ligation and precipitation.
- Transformation and determination of clone size and recombinant rate by PCR.
- Please note that clone array is not included. Transformed cells will be shipped on dry ice. We recommend that cells be arrayed within 3 days following receipt.
- Library must be stored at -80 degrees.
Major advantages of Bio S&T’s ASH technique versus SSH subtraction method:
| ASH breakthrough technology | SSH method | |
|
ASH cDNA subtraction does not use a normalization (self-eliminating) step. |
SSH method has a normalization step which causes self-elimination of differentially-expressed genes. This causes a representation bias. |
|
|
ASH cDNA subtraction is effective for longer cDNA (usually >1.2Kb). cDNA is full-length-enriched and independent of PCR suppression. |
SSH method requires truncated cDNA (usually <700bp) and is dependent on PCR suppression. This reduces the subtraction effect. SSH method is not effective for longer cDNA. |
|
|
ASH cDNA subtraction provides non-exponential amplification before subtraction because double-stranded cDNA is not required. |
SSH typically requires a PCR amplification step prior to hybridization. This causes a representation bias prior to the subtraction step thereby reducing the subtraction effect. |
|
|
ASH cDNA cloning is directional and allows for construction of expression libraries. |
SSH cloning is non-directional. Directional functional screening is not possible. |

